When using PCR in PGD, one is faced with a problem that is inexistent in routine genetic analysis: the minute amounts of available genomic DNA. As PGD is performed on single cells, PCR has to be adapted and pushed to its physical limits, and use the minimum amount of template possible: one strand. This implies a long process of fine-tuning of the PCR conditions and a susceptibility to all the problems of conventional PCR, but several degrees intensified.
There are three main sources of error in PGD that are specifically addressed by our protocol:
1. Contamination: The high number of needed PCR cycles and the limited amount of template makes single-cell PCR very sensitive to contamination. Contamination of the biopsied cell with DNA of paternal, maternal or extra-familial origin can easily lead to misdiagnosis. Therefore stringent experimental practices are needing: equipment and reagents are reserved just for PGD. Extraneous DNA from sperm or maternal cumulus cells are a potential source of contamination from biopsied embryonic cells collected after IVF procedures. Steps taken to prevent contamination include 1) ICSI to introduce single sperm into the cytoplasm of an oocyte. 2) Biopsied cells are washed in droplets of PBS or medium and the wash drop is tested for contamination. ‘Carry over’ contamination of previous PCR products poses the most significant risk. A separate PCR set up room is used and this may be under constant positive pressure to avoid entry of dust and PCR products. Gowns, gloves, overshoes etc. remain in this room, together with all apparatus. One way to reduce the risk of contamination is to use nested PCR because the second round PCR product, which is in excess, can not be amplified by the first round outer primers. Our PGD protocols include testing both of the embryo washes and our reagents to detect possible contamination. In addition to these controls, we include in each assay polymorphic markers to confirm that the DNA being tested is of embryonic origin. The contamination rate (including wash blanks and reagent blanks) is less than 1%. These rates are very similar to those published by the international community.
2. Allele Drop-Out (ADO): Another problem specific to single-cell PCR is the allele drop out (ADO) phenomenon. It consists of the random non-amplification of one of the alleles present in a heterozygous sample. ADO seriously compromises the reliability of PGD as a heterozygous embryo could be diagnosed as affected or unaffected depending on which allele would fail to amplify. This is particularly concerning in PGD for autosomal dominant disorders, where ADO of the affected allele could lead to the transfer of an affected embryo. ADO is responsible for the 3 reported cases of misdiagnosis of CF (Crifo 1994, Harper & Handyside 1994, Verlinsky 1996). Some centres have reported ADO rates as high as 30%. Some groups advocate testing 2 cells from each embryo. The probability of ADO affecting the same allele in both cells in low. Although some cells are not sufficiently advanced on day 3 to allow biopsy of 2 cells. It is thought ADO is caused by sub-optimal PCR conditions and/or incomplete cell lysis. The incidence of ADO varies with cell type and unfortunately it appears to be higher in blastomeres. Multiplex PCR can be employed (Findlay 1995) to screen both the disease causing mutation and an informative linked marker which allows 2 chances to detect the mutant gene. ADO is independent for each fragment in a multiplex PCR, even if the amplified fragments lie within a few 100 base pair of each other. Multiplex PCR therefore reduces the chance of misdiagnosis. To address this issue, we always use a polymorphic marker to confirm that paternal and maternal copies of the gene are present in the biopsied cell.
3. Preferential Amplification (PA): Preferential amplification refers to the favored amplification of one allele over the other in the cell. This presents a problem in genotyping, as it can mask the presence of a heterozygote. The problem of preferential amplification is seen in about 25% of cases. To address this issue, we utilize stringent criteria to detect potential heterozygosity. Fluorescent PCR (F-PCR) offers many advantages for PA reduction. It is a more sensitive technique so both alleles are seen even if there is preferential amplification of one allele over another.
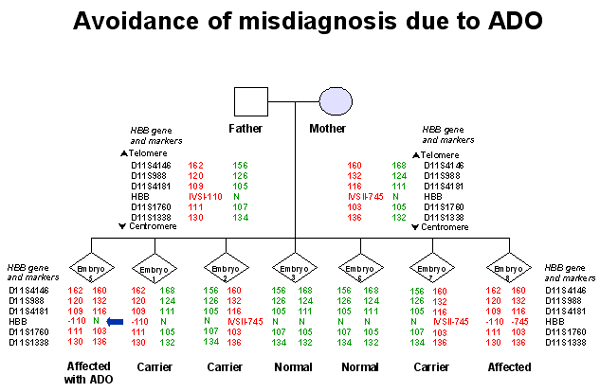
4. Linked STR markers: Including a panel of polymorphic STR markers in the diagnostic assay, closely linked to the gene regions containing the disease causing mutations, increases the robustness of the diagnostic procedure. In fact, determination of the specific STR haplotype associated with the mutation acts both as a diagnostic tool for indirect mutation analysis, providing an additional confirmation of the results obtained with the direct genotyping procedure, and as a control of misdiagnosis due to undetected ADO (Rechitsky et al., 1999)(Figures 1 and 2). The multiplex STR marker system also provides an additional control for contamination with exogenous DNA, as other alleles, differing in size from those of the parents, would be detected (Piyamongkol et al., 2001). The experience of a large series of PGD cycles strongly suggests that PGD protocols for SGD are not appropriate for clinical practice without including a set of linked STR markers, consequently this strategy is currently followed for all our PCR protocols. Diagnosis is assigned only when haplotype profiles, obtained from linked STR markers, and mutation analysis profiles were concordant.