For diseases involving a heterogeneous spectrum of mutations identified, such as Cystic Fibrosis, ?-Thalassemia or Haemophilia A, the development of a mutation-based PGD strategy is not practical because it requires time and resources for standardization of PCR protocols unique for the specific mutations of interest. For these kinds of monogenic diseases, the use of a diagnostic strategy capable of detecting a wide spectrum of mutations and compound genotypes is more feasible.
GENOMA PGD lab conceived and implemented the innovation of using Minisequencing technique for mutation detection on single cells. This procedure is now widely used by most of the centers performing PGD testing.

Minisequencing technique is based on the single dideoxynucleotide extension of unlabelled oligonucleotide primers. The primer extension reaction is performed starting from the purified amplified target. Specific minisequencing primers, which are exactly one base short of the mutation sites, are used for each mutation under investigation. Primers bind to the complementary templates in the presence of fluorescent-labelled ddNTPs and AmpliTaq® DNA Polymerase. The polymerase then adds a single ddNTP at the 3’ end of each primer, complementarily according to the sequence. Since the reaction contains only template, primer, dye-labelled ddNTP and does not include dNTPs, interruption of the reaction occurs after only one incorporation of a dideoxy terminator. This process is repeated in successive rounds of extension and termination, thus the resulting products, varying in colour, can then be analyzed by electrophoresis (Figure 1).
The colours of the final peaks are determined by the specific genotype at this locus, making it possible to identify the base variation. The mutation site can thus be reliably differentiated between homozygous wild type and mutant (one peak of a specific colour) or heterozygous. In the latter case, two differently-coloured peaks occur in the electropherogram, one derived from the normal base and the other from the mutated base.
The minisequencing method involves the use of a fluorescence-based DNA mutation analysis protocol. When compared with conventional methods, fluorescence-based protocols offer many distinct advantages for clinical PGD. Firstly, fluorescence permits PGD sensitivity, as well as accuracy and reliability, to be substantially increased (Findlay et al., 1995 and 1998). Secondly, packaged computer software allows easy data interpretation and management. Furthermore, fluorescence-based systems are highly amenable to multiplexing, which has great potential regarding detection of multiple mutation sites. This strategy can be accurate enough to distinguish different kinds of mutations, such as single nucleotide substitution and small deletions or insertions.
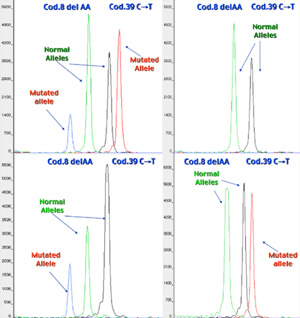
Minisequencing results of a Beta Thalassemia PGD case, in which the mutations of interest were Cod.8 del-AA and Cod
The main benefit of minisequencing is the use of a mutation analysis protocol based on a common procedure for each mutation to be analyzed. This is very important for the PGD of genetic diseases characterized by a heterogeneous spectrum of mutations identified, such as Cystic Fibrosis, B-Thalassemia or Haemophilia A. In fact, for this kind of monogenic diseases, the use of a common procedure able to detect a wide spectrum of mutations, and compound genotypes becomes more feasible. The development of different PGD protocols specific for each mutation to be analysed is thus avoided. With the use of minisequencing for mutation detection, the PGD strategy is the same for the diverse clinical cases involving different mutations: external and nested PCR, purification of PCR products, minisequencing reaction and capillary electrophoresis. The only difference between one PGD case and another that involves PCR amplification of the same DNA region, but with a different mutation panel, lies in the use of different minisequencing primers that can be easily designed and whose application does not require the need of extensive trial testing.
Another useful feature of minisequencing is that different mutation sites can be investigated simultaneously ("multiplexing”), even if they are located in different regions of the gene, depending on the number of primers used in the minisequencing reaction. In multiplex reactions, multiple primer/template combinations are used in a single tube reaction format, and products are analyzed in a single electrophoresis run.